Viral Transcriptomics
Research Contents
Main Purpose
The Viral Transcriptomics Team will elucidate the transcriptomic features and host–virus interactions of cross-species infecting viruses, such as coronaviruses and influenza viruses. Multi-species and multi-organ organoids provide host-specific environments that allow for detailed investigations into viral replication dynamics, cross-species adaptation, and host immune responses. Furthermore, the integration of genomic and (epi)transcriptomic profiling techniques enhances our ability to identify critical determinants of zoonotic transmission.
Our Research Focus (What We Study)
This research aims to leverage organoid platforms and viral (epi)transcriptomics approaches to:
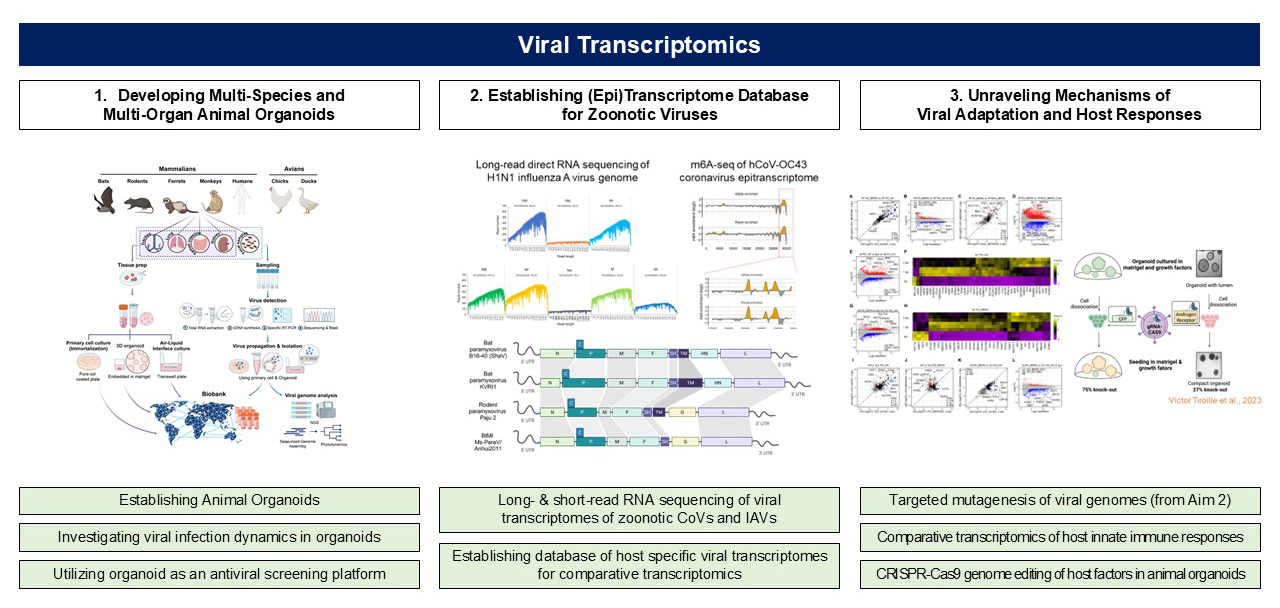
1. Develop multi-species and multi-organ animal organoids for studying zoonotic respiratory viruses
2. Characterize the viral genome and (epi)transcriptomic features that influence species specificity and adaptation
3. Functionally validate key viral and host determinants through genome engineering and transcriptomic analysis
Our Research Approach (How We Study)
- Establishment of Multi-Species, Multi-Organ Animal Organoid Models
- Development of organoids derived from key zoonotic viral hosts, including bats, birds, and various mammalian species
- Analysis of replication and cross-species adaptation of coronaviruses and influenza viruses in a host species- and tissue-specific manner Application of single-organoid immunohistochemistry (so-IHC), spatial transcriptomics, and single-cell transcriptomic analysis
- Establishment of a 2D organoid-based culture platform for antiviral drug screening
- Comparative Transcriptomic Analysis of Zoonotic Viruses
- Construction of host-specific transcriptomic and epitranscriptomic databases for coronaviruses and influenza viruses
- Implementation of short-/long-read RNA sequencing and various sequencing techniques including m6A-seq
- Comparative analysis of host-specific viral transcriptomes to elucidate cross-species adaptation and transmission dynamics
- Functional Characterization of Key Host-Virus Interaction Genes during
- Functional validation of viral genome mutations using recombinant viruses
- Comparative transcriptomic analysis of innate immune responses by virus and host species
- Functional validation of host genes via CRISPR-based genome editing
- Identification and functional analysis of host RNA-binding proteins interacting with viral RNAs using RAP-MS